STRENGTH
Leading to Success
with high expertise and attentive service
Our professional staff will provide you
end-to-end satisfaction.

Building trustful relationships
In research support services, the success of a project cannot be achieved without building a good trust relationship with our customers. There are various situations in which researchers are extremely busy and unable to follow the rapid technological advances in omics analysis. With that in mind, Rhelixa offers support that closely aligns with the needs of researchers.


High expertise
Our company has many researchers on staff, including M.D.s and Ph.D.s. Researchers with a track record of leading large-scale projects in clinical and basic research using omics analysis will support you in experimental design, examination, and bioinformatics. Furthermore, we offer customized and integrated analysis tailored to your research objectives, rather than relying on single-process analysis pipelines in bioinformatics.

Reasonable pricing
Our lab is equipped with the highest level of research facilities, but we deliberately keep the ownership of next-generation sequencers to a minimum. This is because we believe that being tied down to "keeping the equipment running" in the increasingly diverse and complex field of omics research would make it difficult to pursue customer-oriented services that Relixa aims for. As a result, we collaborate with competitive providers both domestically and internationally in the sequencing process, achieving low-cost and high-quality data acquisition.

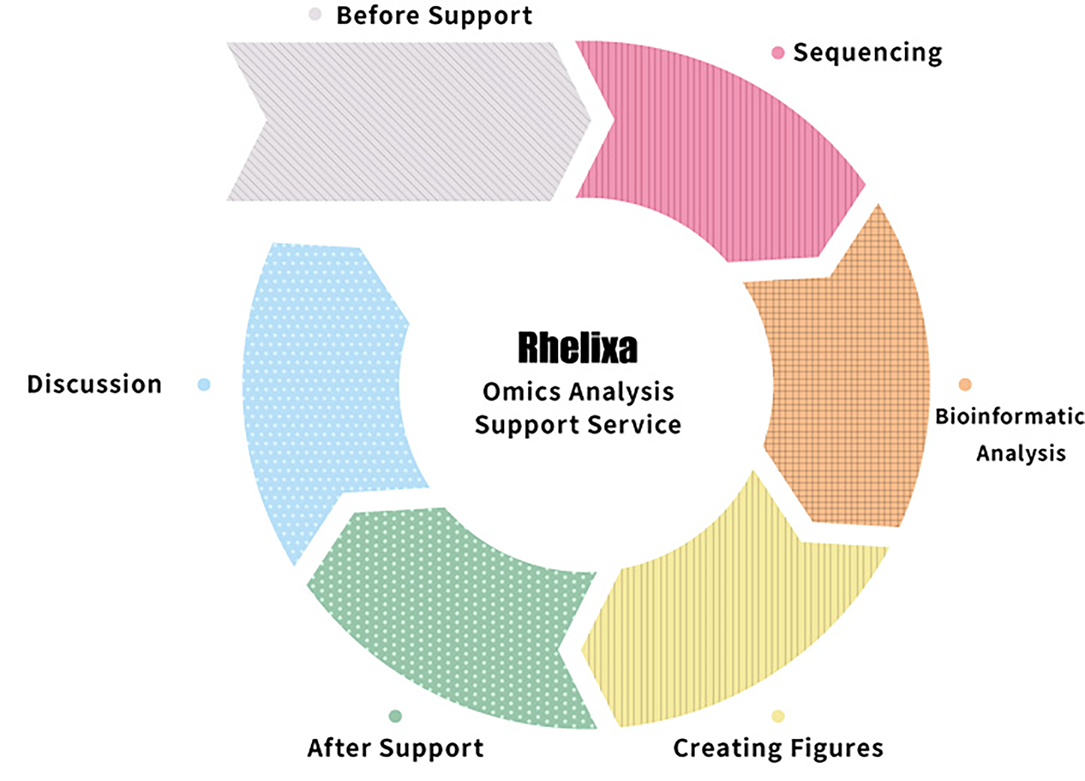
SUPPORTWe support all aspects of your research,
from study design to manuscript preparation and practical application.
Academic research support
Corporate research support

Co-authored Papers
Sci Rep. 2023 Aug 9;13(1):12888. doi: 10.1038/s41598-023-40051-6.
Sci Transl Med. 2023 Jun 14;15(700):eabq7721. doi: 10.1126/scitranslmed.abq7721.
G3 (Bethesda). 2022 Jul 1:jkac166. doi: 10.1093/g3journal/jkac166. Online ahead of print.
Cell Rep. 2022 Feb 8;38(6):110332. doi: 10.1016/j.celrep.2022.110332.
Nat Commun. 2021 Dec 2;12(1):7045. doi: 10.1038/s41467-021-27321-5.
Sci Rep. 2021 Sep 21;11(1):18687. doi: 10.1038/s41598-021-98344-7.
Sci Signal. 2021 Jan 26;14(667):eabb3616. doi: 10.1126/scisignal.abb3616.
EMBO J. 2020 Mar e103949. doi.org/10.15252/embj.2019103949
Cell Rep. 2019 Oct 1;29(1):162-175.e9. doi: 10.1016/j.celrep.2019.08.086.
G3 (Bethesda). 2022 Jul 1:jkac166. doi: 10.1093/g3journal/jkac166. Online ahead of print.
Nat Commun. 2021 Dec 2;12(1):7045. doi: 10.1038/s41467-021-27321-5.
Sci Rep. 2021 Sep 21;11(1):18687. doi: 10.1038/s41598-021-98344-7.
Sci Signal. 2021 Jan 26;14(667):eabb3616. doi: 10.1126/scisignal.abb3616.
EMBO J. 2020 Mar e103949. doi.org/10.15252/embj.2019103949
Cell Rep. 2019 Oct 1;29(1):162-175.e9. doi: 10.1016/j.celrep.2019.08.086.
Epigenetics & Chromatin 12, 77 (2019). doi.org/10.1186/s13072-019-0319-0
EMBO Rep. 2018 Dec;19(12):e46255. doi:10.15252/embr.201846255
EMBO J. 2018 Jul 2;37(13). pii: e97723. doi: 10.15252/embj.201797723.
Nat Commun. 2018 Apr 19;9(1):1566. doi: 10.1038/s41467-018-03868-8.
Sci Rep. 2018 Feb 28;8(1):3779. doi: 10.1038/s41598-018-22180-5.
Nat Cell Biol. 2017 Sep;19(9):1081-1092. doi: 10.1038/ncb3590.
Nucleic Acids Res. 2017 May 5;45(8):4344-4358. doi: 10.1093/nar/gkx159.
Cell Rep. 2017 Feb 28;18(9):2228-2242. doi: 10.1016/j.celrep.2017.02.006.
Mol Cell. 2015 Nov 19;60(4):584-96. doi: 10.1016/j.molcel.2015.10.025.
Nat Commun. 2015 May 7;6:7052. doi: 10.1038/ncomms8052.